1. INTRODUCTION
A. annua L. (Asteraceae) is a widely distributed aromatic herb known for producing artemisinin, a key anti-malarial compound whose discovery earned Tu Youyou the 2015 Nobel Prize. Beyond malaria treatment, the species exhibits broad biological activities, leading to extensive research on its biosynthesis and cultivation optimization (Septembre-Malaterre et al., 2020 [1]; Ferreira, 2007 [2]).
In Vietnam, research since the late 1980s has aimed to establish domestic artemisinin sources, documenting the plant’s chemical profile and distribution (Woerdenbag et al., 1994) [3]. However, morphological similarities with other species like A. apiacea, A. capillaris and A. campestris frequently lead to market adulteration and misidentification. Accurate taxonomic identification is therefore critical to ensure the quality and safety of medicinal materials, avoiding the health risks associated with using incorrect species.
2. MATERIALS AND METHODS
2.1. Materials
Five A. annua samples were collected from Thuận Châu (TC), Sông Mã (SM), Mộc Châu (MC), Bắc Yên (BY) and Quỳnh Nhai (QN) districts of Sơn La Province, Vietnam.
2.2. Methods
DNA extraction and PCR
Total genomic DNA was extracted using the CTAB method. PCR amplification targeted three barcode regions rbcL, matK and 18S, using specific primer pairs. Protocols followed Kress et al. (2009) [4].
Sequence analysis
PCR products were sequenced bidirectionally using the Sanger method with BigDye™ Terminator chemistry on an ABI 3500 system. Sequence editing was performed using ATGC and BioEdit. Quality trimming was applied at Phred >20. BLASTn was used to compare sequences with GenBank references.
Phylogenetic analysis
Sequences were aligned with Clustal Omega. Phylogenetic trees were constructed in MEGA 11 using the Maximum Likelihood method with 100 bootstraps. Genetic distances were calculated using BioEdit.
3. RESULTS AND DISCUSSION
3.1. PCR amplification and sequencing
Total genomic DNA extracted and purified using the CTAB method [5] was amplified for the three target regions rbcL, matK and 18S using specific primer sets. The PCR reactions showed 100% success, producing clear and distinct amplification bands without any nonspecific products. The resulting amplicons were approximately 600 bp, 800 bp and over 1 kb in length, respectively, consistent with theoretical expectations. All PCR products displayed high purity, no detectable contaminants, and a single, strong band, ensuring they were suitable for subsequent sequencing.

Figure 1. PCR electrophoresis results of total genomic DNA from the five samples using primer sets for rbcL (A), matK(B) and 18S(C). Marker 1 kb DNA Ladder (Thermo).
3.2. Sequencing results
The sequences of the rbcL, matK and 18S regions obtained from the five Artemisia annua samples collected in Sơn La were quality-checked using FinchTV (Digital World Biology Products, USA) and only bases with Phred scores higher than 20 were retained for analysis. The sequences were subsequently examined in chromatogram form using BioEdit, where clear and well-defined peaks corresponding to the four nucleotides were observed (Figure 2).
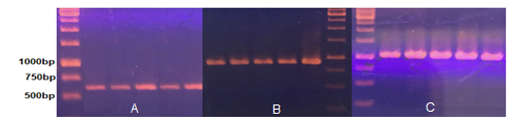
Figure 2. Sequencing chromatograms of the TC sample.
Bidirectional sequencing was performed to obtain complete and accurate sequences. The results indicated that the 18S, rbcL, and matK sequences were 1023 bp, 624 bp, and 837 bp in length, respectively. Consequently, the sizes of the obtained sequences corresponded to the gel electrophoresis results, demonstrating that the sequences were intact and the entire gene regions were identified.
3.3. Species Identification Results
Sequences were aligned using Clustal Omega and queried via BLASTn, revealing 98–99% similarity to A. annua. For phylogenetic analysis, the top 20 sequences (>96% similarity) were selected to construct a Maximum Likelihood tree using MEGA 11 with 100 bootstrap replicates. Results for the matK region are shown in Figure 3.

Figure 3. Phylogenetic tree based on the matK gene sequences
The phylogenetic trees based on matK (Figure 3) and rbcL demonstrated distinct species delineation within the Artemisia genus. The Son La samples formed a stable, monophyletic clade with A. annua reference sequences (e.g., KF056292, KX587981), distinct from other congeneric species like A. selengeensis and A. carvifolia. This confirms the efficacy of matK and rbcL as robust DNA barcodes. In contrast, the 18S tree, while showing high similarity to A. annua, failed to fully resolve species-level relationships due to its high conservation. Consequently, based primarily on the high-resolution matK and rbcL data, the Son La samples are definitively identified as A. annua.
3.4. Genetic Diversity Assessment Results
Sequence variations in the rbcL, matK and 18S regions among the surveyed samples were expressed through similarity coefficients obtained using BioEdit software to measure genetic distance. The results indicated that the nucleotide sequence similarity among the five collected Artemisia annua individuals ranged from 94% to 100%. Based on the nucleotide sequence analysis results, a phylogenetic tree was constructed (Figure 4).

Figure 4. Phylogenetic trees illustrate the genetic diversity of A. annua populations in Son La based on three gene regions: rbcL (A), matK (B) and 18S (C).
Molecular phylogenetic analysis based on the 18S, rbcL and matK gene segments indicated that the five A. annua L. populations collected in Son La exhibit high genetic diversity, reflecting a genetic structure dependent on environmental conditions and geographic location. The branching structures within the trees possessed very high bootstrap values (92–100%), confirming the reliability of the grouping relationships. However, there was inconsistency among markers in determining the closest relationships. Both rbcL and matK markers strongly reflected the grouping of Thuan Chau and Song Ma populations with high bootstrap support, suggesting a close genetic relationship, possibly due to high gene flow between these two regions. Conversely, the 18S marker placed the Quynh Nhai and Thuan Chau groups adjacent to each other, suggesting that the evolutionary history of the 18S gene may differ from that of the other two gene regions. Notably, the Moc Chau population, located in the Southeast, tended to branch off earliest in both the 18S and rbcL trees, indicating it may be the most genetically distinct population among the studied samples.
The chromatograms displayed peaks representing the four nucleotides; sharp, clear, and well-separated peaks in the first 500 bp indicated relatively high sequence quality. However, signals began to blur and overlap in later nucleotides. This phenomenon is a common characteristic of Sanger sequencing (Al-Shuhaib and Hashim, 2023) [6]. This may be caused by reduced chain extension efficiency in later cycles, DNA fragmentation, or differences in migration rates of DNA fragments within the capillary, causing long fragments to separate poorly and generate overlapping signals (Mero, 2021) [7]. To overcome this limitation, bidirectional sequencing is necessary to minimize errors and obtain complete and accurate sequences.
In this study, intraspecific divergence was observed in the rbcL gene region, while significantly low sequence divergence was noted in the matK gene region. One possible reason is that chloroplast genes evolve more slowly than nuclear genes. The conservation of chloroplast DNA sequences ensures comparability across groups. Consequently, matK sequences are often applied to classify taxa at higher levels, such as genus and species. The rbcL sequence has varying copy numbers across different taxes, leading to high persistence of variants within the genome. The rich diversity of rbcL can be utilized in classifying lower-level taxa (genus, species, subspecies) at the population level. Our study confirmed this to a certain extent.
Although the differences between A. annua from different sampling sites were insufficient for division into distinct species or subspecies, they reflect intrinsic variation and DNA polymorphism in A. annua, while providing information for the optimal use and conservation of A. annua genetic resources.
4. CONCLUTION
The three gene regions rbcL, matK and 18S showed that all five A. annua samples collected in Son La matched 99–100% with Artemisia annua L. on GenBank. Phylogenetic trees confirmed that all samples clustered within the same clade as A. annua, with matK and rbcL demonstrating higher species resolution than 18S. The level of intraspecific variation among sampling regions reflects slight genetic differentiation based on geographic conditions. These results provide a reliable basis for the identification and conservation of A. annua genetic resources in Son La.
ACKNOWLEDGEMENTS
The research is funded by Vietnam Academy of Science and Technology under grant number NCVCC08.05/25-25 (No. 2995/QĐ-VHL)
REFERENCES
1. Septembre-Malaterre, A., L'Honoré Gregoire, F., & Remize, F. (2020), Artemisia annua L.: A potential source of molecules with pharmacological interest. Journal of Ethnopharmacology, 254, 112717. 2. Ferreira, J. F. S. (2007), Nutrient deficiency in the production of artemisinin, dihydroartemisinic acid, and artemisinic acid in Artemisia annua L. Industrial Crops and Products, 26(3), 326–336. 3. Woerdenbag, H. J., Pras, N., Chan, N. G., Bang, B. T., Bos, R., van Uden, W., Van, Y. P., Boi, N. V., Batterman, S., & Lugt, C. B. (1994), Artemisinin, related sesquiterpenes, and essential oil of Artemisia annua during a vegetation period in Vietnam. Planta Medica, 60(3), 272–275. 4. Kress, W. J., Wurdack, K. J., Zimmer, E. A., Weigt, L. A., & Janzen, D. H. (2009), DNA barcodes for land plants. Proceedings of the National Academy of Sciences, 106(31), 12794–12797. 5. Saghai-Maroof M. A., Soliman K. M., Jorgensen R. A., Allard R. W. (1984), Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proceedings of the National Academy of Sciences of the United States of America 81 (24), 8014-8018. 6.Al-Shuhaib, M. B. S., & Hashim, H. O. (2023), Standardization of the visual interpretation of Sanger sequencing chromatograms. Journal of Genetic Engineering and Biotechnology, 21(1), 1–9. 7. Mero, W. M. S. (2021), DNA sequencing methods: A review. Academic Journal of Nawroz University, 10(2), 168–174.
Le Thi Bich Thuy, Ton Son Bach, Tran Thi Luong - Institute of Biology, Vietnam Academy of Science and Technology
Nguyễn Văn Huấn - Phenikaa University
Nguyễn Như Toản - Hanoi Metropolian University




































